Proximity Labeling: DEL Selection Without Capturing the Target

DNA-encoded library (DEL) selections typically use affinity pulldown to enrich compounds that bind targets of interest. While it is highly effective at identifying hits for numerous targets, affinity pulldown also has its limitations. For example, protein targets that are captured onto a solid-phase matrix may lose their native structure and activity. Desirable binding sites on the target may also be obscured and matrix-dependent unnatural binding sites could be created, potentially enriching compounds that are not physiologically relevant. Moreover, stringent washing is often required to achieve the desirable signal-to-noise ratio, and the unfortunate side effects of that process could be the skewing of selection output toward slower-off-rate binders and a confounded relationship between enrichment and affinity.
Recently, the Krusemark lab at Purdue University reported a novel in-solution proximity labeling selection approach.1 Instead of affinity-tagging the protein target, the authors took inspiration from the protein-protein interaction (PPI) mapping literature and tagged the target with ultraID, a small enzyme that produces biotin-AMP, a short-lived reactive species.2 As biotin-AMP diffuses from ultraID’s active site, it has a limited lifetime to biotinylate a nucleophile before being quenched by water, leading to a limited “striking distance” of about 10 nm. This behavior of selectively labeling within a close proximity provides a mechanism for target-bound DEL members to be preferentially biotinylated. This, in turn, allows them to be enriched in a subsequent step via streptavidin capture. Importantly, the recording of the ligand-target binding event occurs in solution, thus eliminating the need to capture the target onto a matrix.
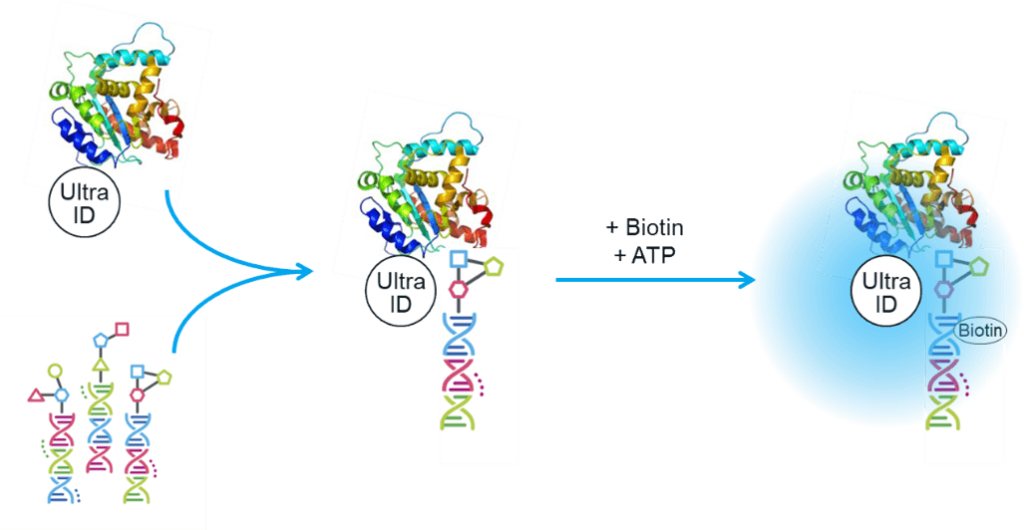
Using single-stranded DNA-linked compounds, each of which were hybridized to an amine-functionalized complement strand (which serves as the nucleophile for biotinylation), the authors demonstrated 6,300-fold enrichment of a 50 nM ligand on ultraID-tagged CBX7 chromodomain, with respect to a nonligand. Crucially, the DNA-linked compound could be:
- Recovered by strand separation
- Hybridized to a fresh amine-functionalized strand
- Subjected to further selection to achieve 500,000-fold overall enrichment (2 rounds)
In a scenario that may be more relevant to de novo hit identification, a weaker binder (Kd ≈ 2 µM to CBX7 chromodomain) demonstrated 400-fold enrichment over one round and 11,000-fold enrichment over two rounds. In another application of their technology, the authors performed a selection against ultraID-tagged δ opioid receptors on the surface of live cells and achieved 280-fold enrichment for a subnanomolar ligand, as well as 40-fold enrichment for a ~10 nM ligand.
In addition to ultraID, the authors developed a second proximity-dependent enrichment system based on the ability of Nluc (a small luciferase enzyme) to unmask coumarin-protected biotin within a limited radius (via bioluminescence resonance energy transfer, or BRET). Although the ligand-to-nonligand enrichment (270-fold with CBX7 chromodomain) was less impressive due to higher background reactivity, the development of this orthogonal method could provide opportunities for multiplexing with the ultraID approach.
Overall, this new paper introduced two innovative additions to the in-solution DEL selection repertoire. These methods previously consisted of interaction-dependent PCR and binder trap enrichment, both of which relied on tagging the target protein with an oligonucleotide. We look forward to seeing these new methods used on a chemically diverse library to enrich novel engagers of high-value targets. You can bet that X-Chem will be closely watching this space!
References
- Cai, B. et al. Selection methods for proximity-dependent enrichment of ligands from DNA-encoded libraries using enzymatic fusion proteins. Chem. Sci. 2023,14, 245-250.
- Kubitz, L. et al. Engineering of ultraID, a compact and hyperactive enzyme for proximity-dependent biotinylation in living cells. Commun Biol. 2022, 5, 657.
Flow Chemistry for Contemporary Isotope Labeling
Isotope-labeled compounds have emerged as important tools in drug discovery and development. The incorporated isotopes, particularly deuterium, in pharmaceuticals, have...
Driving Discovery Through Miniaturized High-Throughput Chemistry
When chemists approach the formation of a new bond, they often choose reaction conditions that they (or their colleagues) are...